 SCIENTIFIC CALCULATOR
SCIENTIFIC CALCULATOR
☞
Let's Help Scientists Develop New Treatments
☜
Do you have a computer at home that sits mostly idle? You can put it to some useful work.
During the COVID pandemic we joined thousands of donors of CPU and GPU computing capacity to help the Folding@Home project. By doing so, our excess computing resources support scientists in simulating and understanding molecular dynamics of proteins. Many of the projects that utilize this distributed computing platform are focusing on COVID-19 and other viruses and proteins in search for therapeutic opportunities.
If you want to connect your computer to this project, you can start by installing the Folding@Home packages on your computer. Then you can follow a few easy configuration steps.
Once you get it up and running and a Work Unit is assigned to your computer, you can visualize the very molecule that is being analyzed...
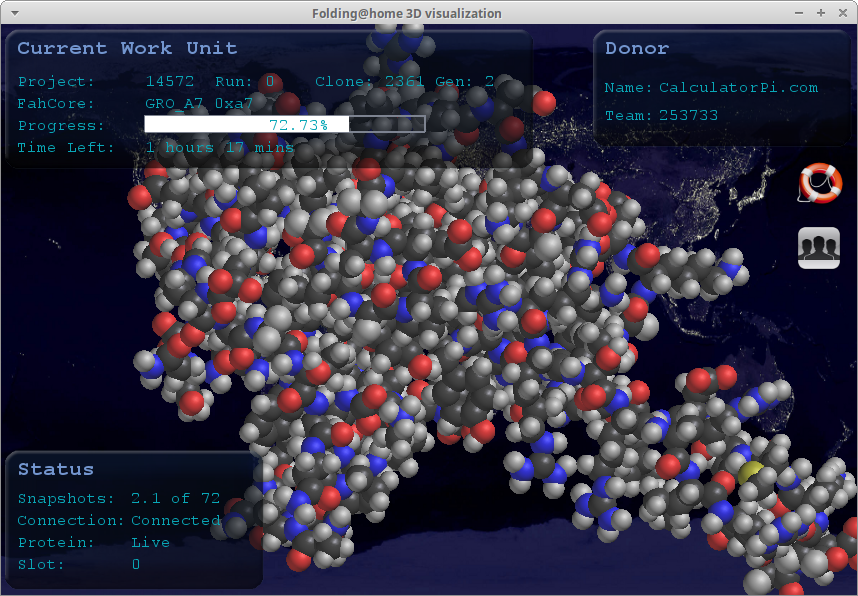
-
This TED talk is a short but exciting introduction to protein molecules.
This video by David Baker from University of Washington nicely explains what are the objectives, challenges, methods and successes of protein folding simulations (beyond FAH).
This paper goes deeper - but not too deep - into what proteins are and why we study them.
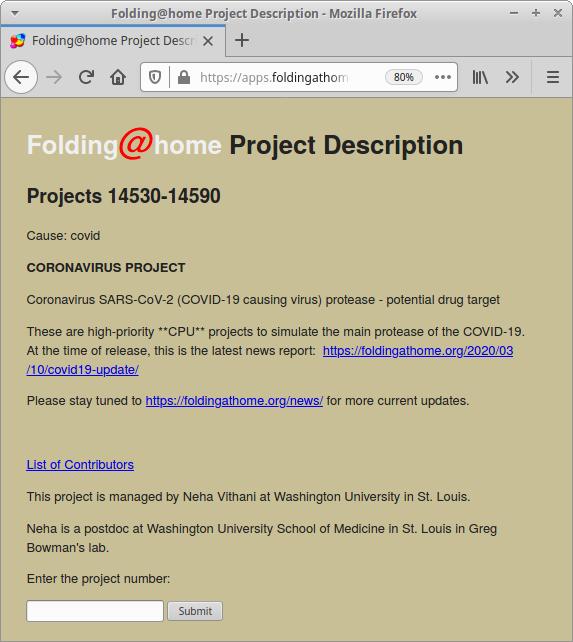
This page lists specific COVID-19 related projects supported by FAH.
You can look up our contribution summary and even join our team # 253733.
A tip: If you worry about security (we really don't in this case) you may consider running Folding@Home inside VirtualBox. You can get it from here.
Pi Day 2021 update: After 1 year of folding, along with our Team members, we are happy to report contributing over 4300 work units to the FAH project.

